Tools & Techniques
A human CRISPR-like system adds a new approach to regulating translation
A human system modeled on CRISPR might address the precision, immunogenicity and delivery challenges in the field of RNA control
A new system modeled on CRISPR and made entirely of human proteins is the latest strategy to intervene in gene control at the RNA translation level. The technology could bring higher levels of precision and lower risks of immunogenicity than other RNA control methods.
The next step is to test it in models of disease.
In a Cell study, a group of University of Chicago researchers engineered a system of programmable effectors dubbed CIRTS (CRISPR/Cas-Inspired RNA Targeting System) that, like CRISPR-Cas, can bring effector proteins such as nucleases to a target nucleic acid site.
But while the most commonly used CRISPR systems target DNA sequences, CIRTS is designed to bind RNA sites to make reversible changes in protein expression.
Hitting RNA is emerging as an important mechanism to expand the druggable target space and reach targets that can't be touched by traditional small molecules and biologics.
"In a post-Cas9 world, monogenic diseases will probably be cured. I'm most excited about trying to use this technology to target multigenic diseases."
At least three companies -- Arrakis Therapeutics Inc., Expansion Therapeutics Inc. and Ribometrix Inc. -- have systematic techniques for identifying RNA-binding small molecules. Another company, Anima Biotech Inc., is controlling RNA translation with small molecules that bind translational control elements (see “Anima Opens New Target Space”).
University of Chicago professor Bryan Dickinson, who led the study, said CIRTS should be more precise than small molecules.
In addition, he said, CIRTS uses only human proteins, which decreases the risk of immunogenicity.
“The immune response is really key, especially when targeting RNA. With DNA therapy, you're giving it once. If you get sick and have an immune response, it's unlikely to reoccur. RNA-targeting therapies will be given chronically, more like traditional small molecules and biologic drugs, so an immune reaction is a bigger problem," Dickinson told BioCentury.
CIRTS is also smaller than standard bacterial CRISPR systems, and can be more easily packaged for delivery.
Dickinson and colleagues modeled it on CRISPR-Cas13, one of the more recently identified programmable bacterial CRISPR systems identified that is useful for diagnostic applications (see "A Cutting Diagnosis").
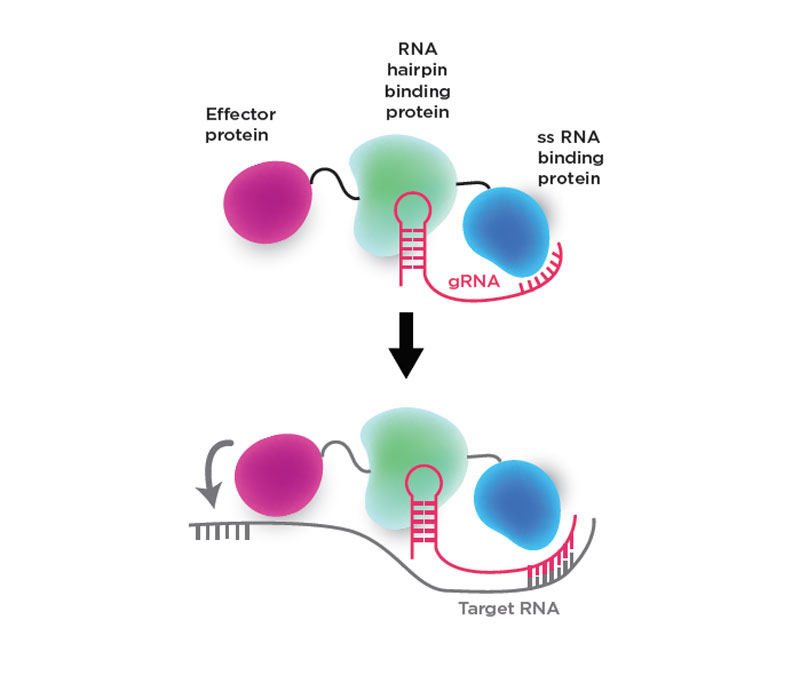
Figure: Components of the CIRTS programmable effectors
By taking bacterial proteins out of the equation, a University of Chicago team thinks they've found a way to make a CRISPR-like system that is less immunogenic. They've also designed it to control translation by delivering different effectors, not just nucleases, to RNA target sites.
In a Cell paper, the group used CRISPR-Cas13 as a model system to design a four-part programmable effector complex called CRISPR-Inspired RNA Targeting System (CIRTS) made from human proteins.
Top: The design includes a guide RNA (gRNA) that directs the proteins to a complementary target RNA sequence, an effector protein such as a nuclease or RNA reader that increases or decreases protein expression, an RNA hairpin binding protein that binds the guide RNA's hairpin structure, and a charged single-stranded RNA binding protein that stabilizes the guide.
Bottom: In the presence of the target RNA sequence, the guide strand binds the complementary region of the RNA, enabling the effector protein to act near the binding site.
Cas13 naturally binds RNA and doesn't employ complex mechanisms to unwind nucleic acid chains to access binding sites. Instead, it binds at sites that are already accessible. According to Dickinson, that shouldn't limit the potential targets because there are plenty of different binding sites within each mRNA.
In experiments, efficiency varied by site and was generally lower than that of Cas13. But, while the CIRTS system was able to bind its target site if single mismatches were introduced, it was more sensitive to mismatches than Cas13b.
Dickinson said that’s not a concern for him at this point.
"Everyone is obsessed with selectivity right now, and there are engineering strategies to make it happen, but for us, showing in vivo efficacy is a bigger current goal," he said. "At the RNA level, an argument you could make is that selectivity or off targets will be less critical than in DNA because it is reversible."
His group also used the CIRTS system to hit multiple targets in the cell line by delivering multiple guides with the same effector or using two CIRTS systems at once.
"In a post-Cas9 world, monogenic diseases will probably be cured. I'm most excited about trying to use this technology to target multigenic diseases, where manipulating DNA at more than one site would be very difficult, and you'd want to do that non-permanently," said Dickinson.
One interesting disease application is wound healing, he said, where increasing levels of an oncogene would be very effective, but you wouldn't want to do that by adding transgenes due to the cancer risk. "Maybe activating genes involved in cell proliferation and repair at an RNA level, transiently, would be a better option," he said.
Dickinson said the concept originated as a research tool to better understand RNA control, and digging further into that complexity could lead to additional types of effectors and targets. "RNA is regulated in so many diverse ways. There are so many nodes and knobs you can turn at the RNA level, and the complexity is just beginning to be understood. We have an opportunity to figure that out."
To create the system, Dickinson’s group used human proteins to mimic the four core components of the Cas13 system: an RNA hairpin-binding protein that interacts with the guide sequence, a guide RNA that interacts with the hairpin binding protein and a target RNA site, a charged protein that binds and stabilizes the guide, and an effector protein that acts on the target site.
Instead of delivering a nuclease to the target site like traditional CRISPR systems, the humanized system was engineered in the study to deliver an adenosine deaminase that makes base edits, a reader protein that recruits translational machinery to increase protein expression and a different reader that recruits demethylation machinery to degrade the target. It could also be used to deliver a nuclease, said Dickinson.
In a human cell line, the CIRTS complex corrected a G to A mutation engineered into a luciferase gene using the deaminase, increased expression of a target protein using the reader that promotes translation and degraded a target mRNA by hitting different binding sites along the gene, but efficiency varied by site.
The team’s next steps include studying the system in cell culture and mouse models of diseases.
Dickinson told BioCentury he's exploring several different options for the technology, including licensing it and using the platform as a starting point for a company.
Companies and institutions
Anima Biotech Inc., Bernardsville, N.J.
Arrakis Therapeutics Inc., Waltham, Mass.
Expansion Therapeutics Inc., San Diego, Calif.
Ribometrix Inc., Durham, N.C.
University of Chicago, Chicago, Ill.
BCIQ Company Profiles